Edinburgh, Scotland
2023 Vallee Scholars Meeting
Vallee Scholars get together as a small group every other year to talk about their research and exchange ideas. In an informal setting, they meet colleagues in different fields of the biomedical sciences and from different institutions, both national and international. This year’s meeting brought those appointed since 2019 to Edinburgh, Scotland, for a two-day meeting in June at which eleven Scholars presented their research after a keynote lecture given by Professor Dame Carol Robinson.
Check out the pictures in the photo gallery to the left!
As well as being a world-renowned chemist, Director of the Kavli Institute and Dr Lee’s Professor of Physical and Theoretical Chemistry at the University of Oxford, and a Vallee Visiting Professor, Dr Robinson is also a Director of the Vallee Foundation. She is a former president of the Royal Society of Chemistry and has received several honorary degrees and numerous awards. She opened the 2023 Vallee Scholars Meeting in the beautifully frescoed Castle Suite at the Caledonian Hotel, Edinburgh, with her keynote lecture on the promises and pitfalls of native mass spectrometry. Her talk charted the evolution from recombinant complexes in detergent micelles to receptor signaling across native membranes. While challenges remain, this field of structural biology in the gas phase provides new opportunities to uncover mechanistic insight on a range of receptors, transporters and membrane embedded enzymes. Her informative lecture and the lively Q&A session that followed it presaged a day and a half of fascinating talks.
Pontus Skoglund, PhD, (Francis Crick Institute) presented recent research on ancient genomics and human evolution. Analyses of ancient human genomes by his lab has revealed natural selection on immunity genes in the Stone Age. He then moved on to some exciting discoveries the Skoglund Lab has made by sequencing 4,000-year-old Yersinia pestis, which causes the plague. They used samples taken from the skeletal remains in two archeological sites in Britain, one in Somerset in the south west, and the other in Cumbria, near the Scottish border. Together, Pontus hopes these studies will help us to understand how human and bacterial genomes evolved in response to one another.
In her talk entitled The Origin and Evolution of De Novo Genes, Li Zhao, PhD, (The Rockefeller University) revealed that, in addition to those genes derived from duplication-related processes, de novo genes, which are genes born from ancestrally non-genic sequences, also contribute to gene and functional innovation. She and her lab have combined base-level whole-genome alignments, computational structure modeling, and functional approaches to study the origination, evolution, and protein structure of lineage-specific de novo genes. They observed a gradual shift in sequence composition, evolutionary rates, and expression patterns based on gene ages. Intriguingly, there were minimal protein structural changes in young and old de novo genes. Ancestral sequence reconstruction revealed that well-folded proteins are often born folded, and many are enriched with transmembrane and signal proteins. Single-cell RNA sequencing data show that these genes are enriched in germ cells, supporting a function and selective force related to reproduction.
Lisa Olshansky, PhD, (University of Illinois) discussed her work behind detecting changes in glutamine concentration in vivo in her presentation: Engineering a Redox-Switchable Artificial Metalloprotein (swArM). With glutamine serving as a hallmark for cancer metabolism, these changes will provide a powerful new way to diagnose pre-cancerous states at the earliest onset of disease. To do this, she and her team are creating artificial metalloproteins that undergo a conformational change upon glutamine binding. In this way, they are able to leverage the energetic impacts of conformational control over metallocofactor reactivity. Building on these findings, work is underway to engineer swArMs in which allosteric glutamine binding triggers a change in the magnetic properties of an embedded metallocofactor.
Madeline Lancaster, PhD, (MRC Laboratory of Molecular Biology) introduced us to the 3D cerebral organoids she and her team have generated from pluripotent stem cells through neural differentiation and a supportive 3D microenvironment. She showed how these organoids, which have the same tissue architecture as the early human fetal brain, are beginning to unravel the mechanisms of human brain size determination, provide insight into regulation of brain size and neuron number across ape species, and highlight key extrinsic regulators of brain development and tissue morphogenesis. Their findings are pointing to key, human-specific aspects of brain development and function that have important implications for neurological disease.
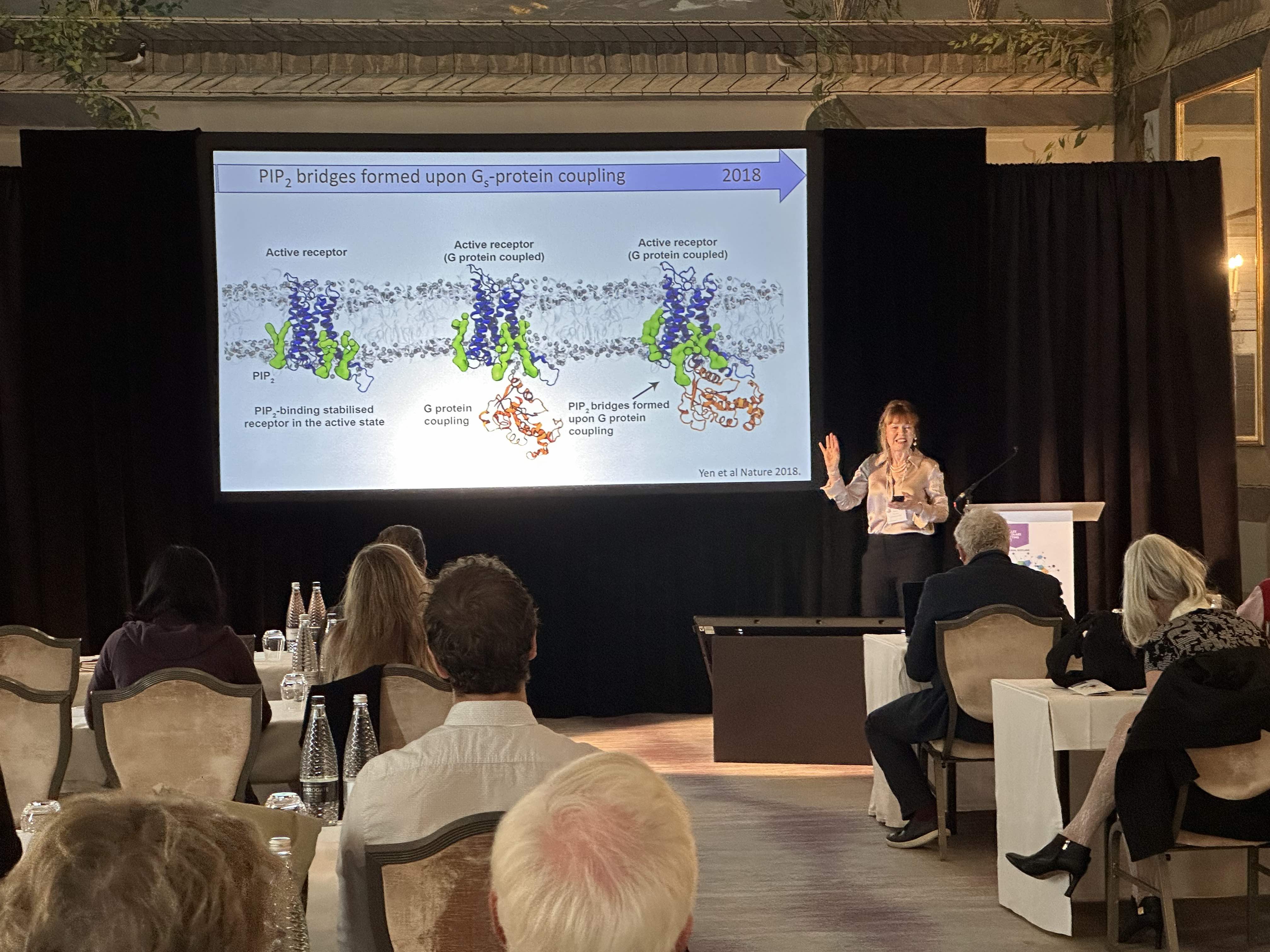
Aashish Manglik, MD, PhD, (University of California, San Francisco), linked back to Dr Robinson’s remarks in his talk focused on G protein-coupled receptors (GPCRs). His collaboration with Dr. Robinson focuses on uncovering the function of poorly understood orphan GPCRs. The main focus of his talk was on visualizing the atomic basis of our sense of smell. Approximately 400 odorant GPCRs encoded in our genome enable us to detect and discriminate the vast diversity of potential odorants. The fundamental molecular logic of how these receptors recognize such a diverse set of odorants to give rise to our perception of smell, however, has remained mysterious. The Manglik lab is trying to decode fundamental principles of odorant recognition by odorant receptors. Analogous to our deep understanding of the visual sensory system, a central hope is that a foundational understanding of odorant molecular recognition may eventually enable precise prediction of an odor precept from the chemical structure of an odorant.
Memories of past events can last, but most molecules activated by learning rapidly decay, as Johannes Gräff, PhE, ( EPFL Lausanne) explained. DNA is one molecular component resistant to turnover. Johannes showed how cell type-specific CRISPR-dCas9-mediated epigenetic editing of a single locus can bidirectionally change memory expression. Such editing is capable of altering memory retention after learning, after having recalled a memory, and is independent of learning-induced neuronal activity. The use of an anti-CRISPR protein showed that such epigenetic editing is potentially reversible, indicating that the observed memory changes are a direct consequence of the epigenetic alterations.
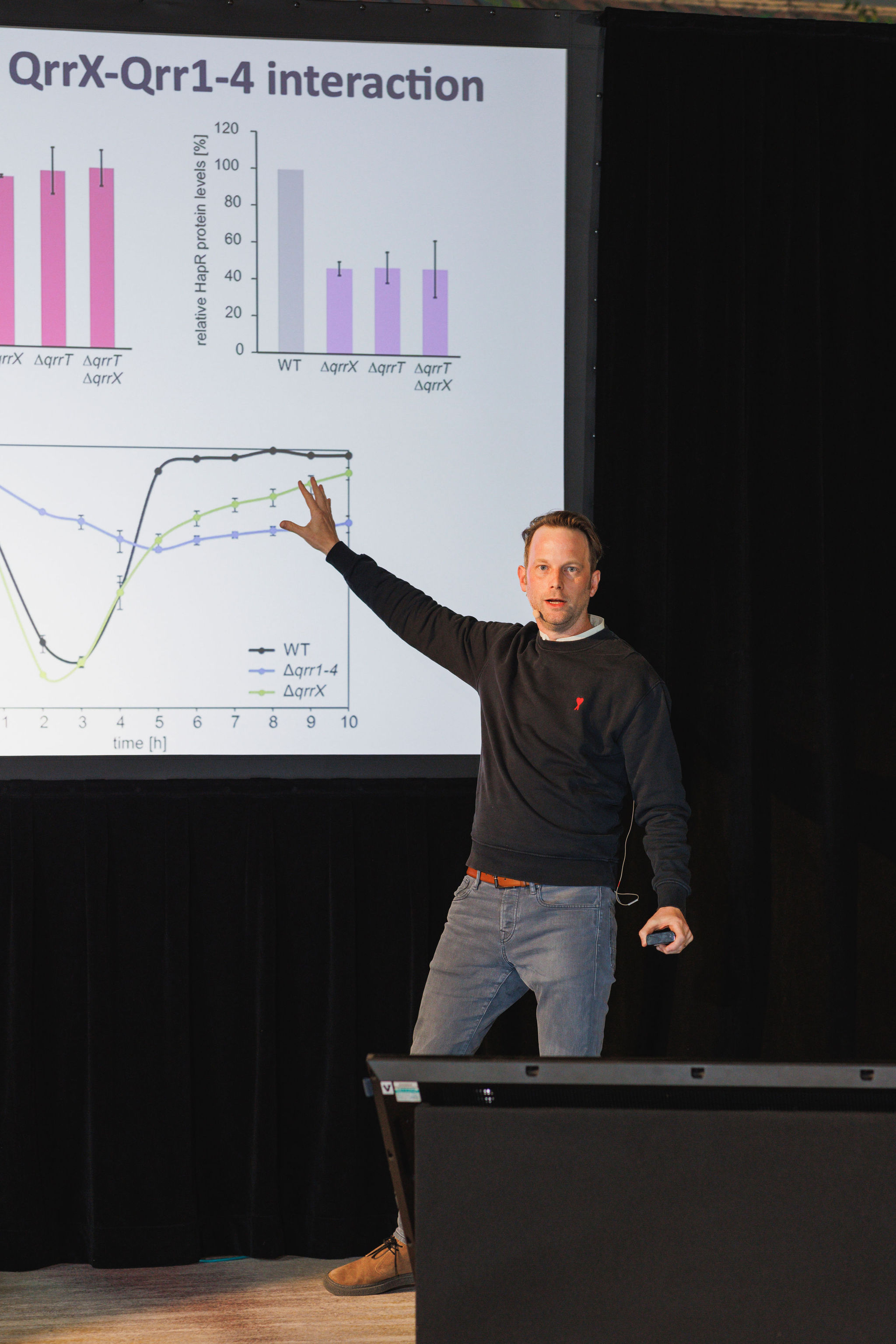
Regulatory RNAs control complex biological functions ranging from embryonic development to bacterial virulence. In bacteria, small RNAs (sRNAs) typically control gene expression by base-pairing with multiple trans-encoded target mRNAs rendering transcript stability and translation initiation. sRNA regulators are modular, versatile, and highly programmable, and therefore have gathered momentum as control devices in synthetic biology and biotechnology. In his presentation, Kai Papenfort, PhD, (University of Jena) highlighted the mechanistic underpinnings of target recognition by bacterial sRNAs and linked these regulatory events to the molecular functions that determine microbial physiology. He also discussed how to harness synthetic regulatory RNAs to decipher microbial phenotypes and the processes underlying sRNA evolution.
Jonathan Abraham, MD, PhD, (Harvard Medical School) reported that mammalian and mosquito orthologs of very low-density lipoprotein receptor are functional receptors for divergent alphaviruses that can cause brain infections in humans and horses. Alphaviruses, to infect cells, must first attach to specific cellular receptors. Alphaviruses can infect numerous vertebrate species and insect vectors separated by hundreds of millions of years of evolutionary history. Interested in how alphaviruses interact with cellular receptors, he and his team used a CRISPR-Cas9-based genetic screen, cell-based assays, and cryo-electron microscopy to further their research. With these tools, he showed that the ability of these alphaviruses to infect a wide range of hosts is a result of their engagement of evolutionarily conserved lipoprotein receptors and that this property contributes to their ability to cause brain infections.lipoprotein receptors and that this property contributes to their neuropathogenesis.
Julian Stingele, PhD, (Ludwig-Maximilians-University, Munich) presented a translation-coupled quality control mechanism that resolves covalent RNA-protein crosslinks. Reactive aldehydes are abundant cytotoxic metabolites which challenge homoeostasis by crosslinking cellular macromolecules. Aldehyde-induced DNA-DNA crosslinks cause cancer and bone marrow failure in Fanconi anemia while DNA-protein crosslinks require proteolytic repair to prevent liver tumors and premature aging. Using photoactivatable ribonucleoside-enhanced crosslinking as a tractable model system to study aldehyde-mimicking RNA damage in the absence of confounding DNA damage, Julian’s team identified RNA damage and RNA-protein crosslink formation as additional central components of aldehyde-induced stress. This lays the groundwork for further research into the cellular responses to these threats.
Ricardo Mallarino, PhD, (Princeton University) spoke about his recent research using the marsupial sugar glider to uncover the molecular basis of the mammalian gliding membrane, or patagium. The sugar glider’s patagium develops postnatally, allowing him to study this structure as it forms and expands. Moreover, sugar gliders belong to a clade of closely related species whose members have independently evolved patagia at least three times, enabling powerful comparative studies. Lastly, sugar gliders complete much of their physical development ex utero, inside of their mother’s pouch. This provides unparalleled access for experimental manipulation and development of molecular tools. Thus, by harnessing naturally evolved phenotypic variation in a species that is amenable to captive breeding/experimental manipulation, Ricardo and his group are uncovering the underlying developmental processes that link genotype and phenotype. More broadly, this work will address how phenotypic novelty is generated at the molecular level - a major long-standing question in the field of evolutionary developmental biology.
Our last speaker, Weizhe Hong, PhD, (UCLA), talked about his research into the neural circuitry that regulates comforting behavior. The ability to help and care for others fosters social cohesiveness and is vital to our physical and emotional well-being. Among the wide diversity of social species, humans and other animals display comforting behavior towards others in distress. To date, no brain area that directly promotes prosocial comforting behavior has been identified. Weizhe demonstrated that mice can display comforting behavior in the form of allogrooming towards their distressed partners. For the first time, this finding establishes the phenomenon of comforting behavior in mice and opens up a new opportunity to study its underlying neural mechanisms. Building upon this discovery, he showed that comforting behavior is encoded by unique patterns of neural activity in a brain region called the medial amygdala. A select subset of neurons in this brain region—tachykinin-expressing GABAergic neurons—directly promotes this behavior.
The lively Q&A sessions spilled over to conversations over the coffee breaks, meals, walks, and quiet moments in the Castle Suite Lobby where Vallee Scholars could be found at all hours in groups of two or three huddled over a laptop. Edinburgh itself was a spectacular location for the meeting. Walking tours of the Old and New Towns gave an overview of the city’s history, architecture, and notable citizens while the Royal Botanic Gardens provided a beautiful setting for dinner one evening. We were een piped into dinner by a stoic bagpiper on the second night. Meeting feedback after the event included: “Unexpectedly stimulating and fun;” “It was the perfect combination of learning, fun people, good food, and beautiful places;” “It really is so energizing to meet with the Vallee community!” and “A phenomenal meeting.” All characteristics that Bert Vallee felt were important to generate good science!